The camp_core_t structure and associated subroutines. More...
Data Types | |
| interface | camp_core_t |
| Part-MC model data. More... | |
Functions/Subroutines | |
| type(camp_core_t) function, pointer | constructor (input_file_path, n_cells) |
| Constructor for camp_core_t. More... | |
| subroutine | load_files (this, input_file_path) |
| Load a set of model data files. More... | |
| subroutine | load (this, input_file_path) |
| Load model data from input files. More... | |
| subroutine | initialize (this) |
| Initialize the model data. More... | |
| logical function | is_initialized (this) |
| Inidicate whether the core has been initialized. More... | |
| logical function | is_solver_initialized (this) |
| Inidicate whether the solver has been initialized. More... | |
| logical function | get_aero_phase (this, aero_phase_name, aero_phase) |
| Get a pointer to an aerosol phase by name. More... | |
| logical function | get_aero_rep (this, aero_rep_name, aero_rep) |
| Get a pointer to an aerosol representation by name. More... | |
| logical function | get_chem_spec_data (this, chem_spec_data) |
| Get a pointer to the chemical species data. More... | |
| logical function | get_mechanism (this, mech_name, mechanism) |
| Get a pointer to a mechanism by name. More... | |
| logical function | get_sub_model (this, sub_model_name, sub_model) |
| Find an sub-model by name. More... | |
| real(kind=dp) function | get_rel_tol (this) |
| Get the relative tolerance for the solver. More... | |
| real(kind=dp) function | get_abs_tol (this, spec_id) |
| Get the absolute tolerance for a species on the state array. More... | |
| type(camp_state_t) function, pointer | new_state_multi_cell (this, env_states) |
| Get a model state variable based on the this set of model data. More... | |
| type(camp_state_t) function, pointer | new_state_one_cell (this, env_state) |
| Get a model state variable based on the this set of model data This is also called for multi-cell systems when no env_state_t array is passed. More... | |
| integer(kind=i_kind) function | state_size (this) |
| Get the size of the state array. More... | |
| integer(kind=i_kind) function | state_size_per_cell (this) |
| Get the size of the state array for each grid cell. More... | |
| type(string_t) function, dimension(:), allocatable | unique_names (this) |
| Get an array of unique names for all species on the state array. More... | |
| logical function | spec_state_id (this, spec_name, state_id) |
| Get the id of a species on the state array by its unique name. More... | |
| subroutine | solver_initialize (this) |
| Initialize the solver. More... | |
| subroutine | free_solver (this) |
| Free the solver memory. More... | |
| subroutine | initialize_aero_rep_update_object (this, aero_rep, update_data) |
| Initialize an update data object for an aerosol representation. More... | |
| subroutine | initialize_rxn_update_object (this, rxn, update_data) |
| Initialize an update data object for a reaction. More... | |
| subroutine | initialize_sub_model_update_object (this, sub_model, update_data) |
| Initialize an update data object for a sub model. More... | |
| subroutine | aero_rep_update_data (this, update_data) |
| Update data associated with an aerosol representation. This function should be called by an external aerosol microphysics model whenever the aerosol condensed data needs updated based on changes in, e.g., particle size or number concentration. The update types are aerosol- representation specific. More... | |
| subroutine | rxn_update_data (this, update_data) |
| Update data associated with a reaction. This function should be called when reaction parameters need updated from the host model. For example, this function can be called to update photolysis rates from a host model's photolysis module. More... | |
| subroutine | sub_model_update_data (this, update_data) |
| Update data associated with a sub-model. This function should be called when sub-model parameters need updated from the host model. More... | |
| subroutine | solve (this, camp_state, time_step, rxn_phase, solver_stats) |
| Integrate the chemical mechanism. More... | |
| integer(kind=i_kind) function | pack_size (this, comm) |
| Determine the size of a binary required to pack the mechanism. More... | |
| subroutine | bin_pack (this, buffer, pos, comm) |
| Pack the given value to the buffer, advancing position. More... | |
| subroutine | bin_unpack (this, buffer, pos, comm) |
| Unpack the given value from the buffer, advancing position. More... | |
| subroutine | do_print (this, file_unit, solver_data_only) |
| Print the core data. More... | |
| subroutine | finalize (this) |
| Finalize the core. More... | |
| subroutine | finalize_array (this) |
| Finalize an array of core data. More... | |
| subroutine | add_aero_phase (this, phase_name) |
| Add a aerosol phase to the model data. More... | |
| subroutine | add_aero_rep (this, rep_name) |
| Add a aerosol representation to the model data. More... | |
| subroutine | add_mechanism (this, mech_name) |
| Add a chemical mechanism to the model data. More... | |
| subroutine | add_sub_model (this, sub_model_name) |
| Add a sub-model to the model data. More... | |
Detailed Description
The camp_core_t structure and associated subroutines.
Function/Subroutine Documentation
◆ add_aero_phase()
|
private |
Add a aerosol phase to the model data.
- Parameters
-
[in,out] this Model data [in] phase_name Aerosol phase name
Definition at line 1763 of file camp_core.F90.
◆ add_aero_rep()
|
private |
Add a aerosol representation to the model data.
- Parameters
-
[in,out] this Model data [in] rep_name Aerosol representation name
Definition at line 1787 of file camp_core.F90.
◆ add_mechanism()
|
private |
Add a chemical mechanism to the model data.
- Parameters
-
[in,out] this Model data [in] mech_name Mechanism name
Definition at line 1813 of file camp_core.F90.
◆ add_sub_model()
|
private |
Add a sub-model to the model data.
- Parameters
-
[in,out] this Model data [in] sub_model_name Sub model name
Definition at line 1838 of file camp_core.F90.
◆ aero_rep_update_data()
|
private |
Update data associated with an aerosol representation. This function should be called by an external aerosol microphysics model whenever the aerosol condensed data needs updated based on changes in, e.g., particle size or number concentration. The update types are aerosol- representation specific.
- Parameters
-
[in] this Chemical model [in] update_data Update data
Definition at line 1291 of file camp_core.F90.
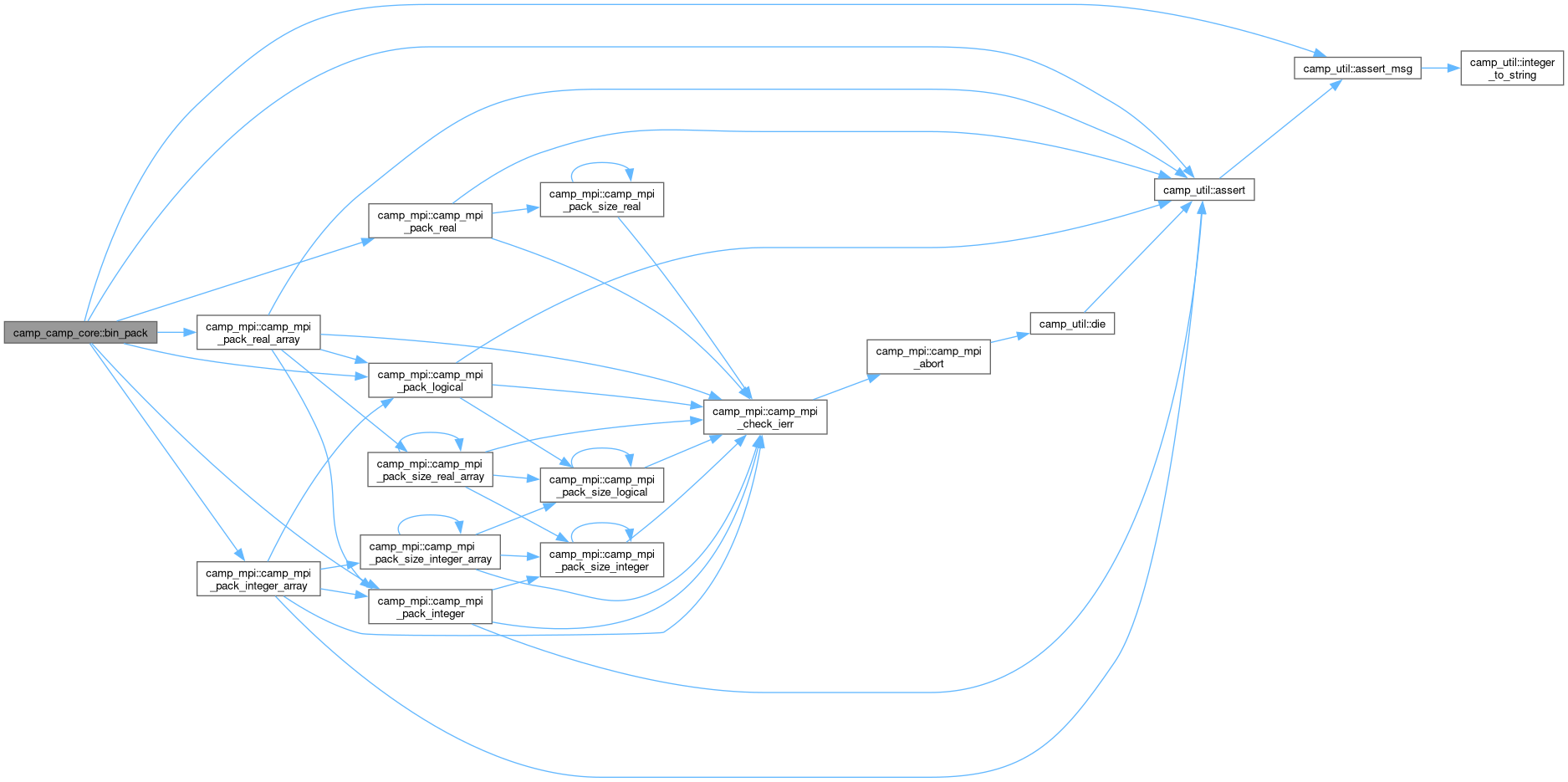
◆ bin_pack()
|
private |
Pack the given value to the buffer, advancing position.
- Parameters
-
[in] this Chemical model [in,out] buffer Memory buffer [in,out] pos Current buffer position [in] comm MPI communicator
Definition at line 1476 of file camp_core.F90.
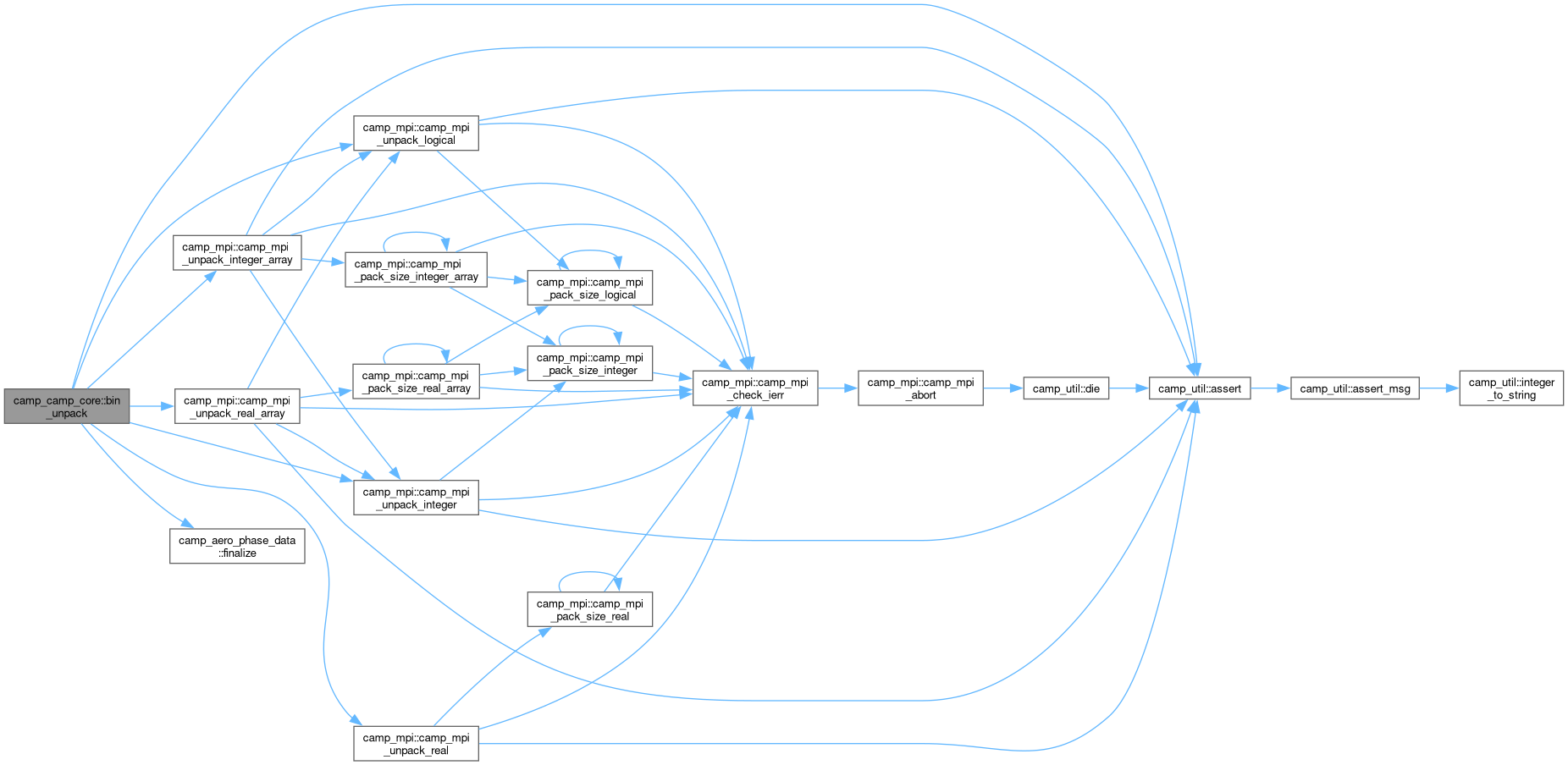
◆ bin_unpack()
|
private |
Unpack the given value from the buffer, advancing position.
- Parameters
-
[in,out] this Chemical model [in,out] buffer Memory buffer [in,out] pos Current buffer position [in] comm MPI communicator
Definition at line 1541 of file camp_core.F90.
◆ constructor()
|
private |
Constructor for camp_core_t.
- Returns
- A new set of model parameters
- Parameters
-
[in] input_file_path Part-MC input file paths n_cells Num cells to compute simulatenously
Definition at line 251 of file camp_core.F90.
◆ do_print()
|
private |
Print the core data.
- Parameters
-
[in] this Core data [in] file_unit File unit for output [in] solver_data_only Print only the solver data (can be used during runtime for debugging)
Definition at line 1609 of file camp_core.F90.

◆ finalize()
|
private |
Finalize the core.
- Parameters
-
[in,out] this CAMP-core data
Definition at line 1716 of file camp_core.F90.
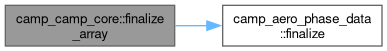
◆ finalize_array()
|
private |
Finalize an array of core data.
- Parameters
-
[in,out] this CAMP-core data
Definition at line 1747 of file camp_core.F90.
◆ free_solver()
|
private |
Free the solver memory.
- Parameters
-
[in,out] this CAMP-core
Definition at line 1207 of file camp_core.F90.
◆ get_abs_tol()
|
private |
Get the absolute tolerance for a species on the state array.
- Returns
- Absolute tolerance
- Parameters
-
[in] this Model data [in] spec_id Species id
Definition at line 962 of file camp_core.F90.


◆ get_aero_phase()
|
private |
Get a pointer to an aerosol phase by name.
- Parameters
-
[in] this Model data [in] aero_phase_name Aerosol phase name to search for [out] aero_phase Pointer to the aerosol phase
Definition at line 822 of file camp_core.F90.
◆ get_aero_rep()
|
private |
Get a pointer to an aerosol representation by name.
- Parameters
-
[in] this Model data [in] aero_rep_name Aerosol representation name to search for [out] aero_rep Aerosol representation
Definition at line 850 of file camp_core.F90.
◆ get_chem_spec_data()
|
private |
Get a pointer to the chemical species data.
- Parameters
-
[in] this Model data chem_spec_data Pointer to the chemical species data
Definition at line 877 of file camp_core.F90.
◆ get_mechanism()
|
private |
Get a pointer to a mechanism by name.
- Parameters
-
[in] this Model data [in] mech_name Mechanism name to search for [out] mechanism Pointer to the mechanism
Definition at line 893 of file camp_core.F90.
◆ get_rel_tol()
|
private |
Get the relative tolerance for the solver.
- Returns
- Relative tolerance
- Parameters
-
[in] this Model data
Definition at line 948 of file camp_core.F90.


◆ get_sub_model()
|
private |
Find an sub-model by name.
- Parameters
-
[in] this Model data [in] sub_model_name Sub model name to search for [out] sub_model Sub model
Definition at line 920 of file camp_core.F90.
◆ initialize()
|
private |
Initialize the model data.
- Parameters
-
[in,out] this Model data
Definition at line 653 of file camp_core.F90.

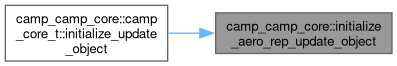
◆ initialize_aero_rep_update_object()
|
private |
Initialize an update data object for an aerosol representation.
- Parameters
-
[in] this CAMP core [in,out] aero_rep Aerosol representation to be updated [out] update_data Update data object
Definition at line 1224 of file camp_core.F90.

◆ initialize_rxn_update_object()
|
private |
Initialize an update data object for a reaction.
- Parameters
-
[in] this CAMP core [in,out] rxn Reaction to be updated [out] update_data Update data object
Definition at line 1245 of file camp_core.F90.

◆ initialize_sub_model_update_object()
|
private |
Initialize an update data object for a sub model.
- Parameters
-
[in] this CAMP core [in,out] sub_model Sub model to be updated [out] update_data Update data object
Definition at line 1266 of file camp_core.F90.

◆ is_initialized()
| logical function camp_camp_core::is_initialized | ( | class(camp_core_t), intent(in) | this | ) |
Inidicate whether the core has been initialized.
- Parameters
-
[in] this Model data
Definition at line 798 of file camp_core.F90.


◆ is_solver_initialized()
|
private |
Inidicate whether the solver has been initialized.
- Parameters
-
[in] this Model data
Definition at line 810 of file camp_core.F90.


◆ load()
|
private |
Load model data from input files.
See Input File Format: CAMP-Chem Configuration Data for the input file format.
- Parameters
-
[in,out] this Model data [in] input_file_path Part-MC input file paths
Definition at line 422 of file camp_core.F90.
◆ load_files()
|
private |
Load a set of model data files.
See Input File Format: CAMP-Chem Configuration File List for the input file format.
- Parameters
-
[in,out] this Model data [in] input_file_path Part-MC input file paths
Definition at line 301 of file camp_core.F90.
◆ new_state_multi_cell()
|
private |
Get a model state variable based on the this set of model data.
- Returns
- New model state
- Parameters
-
[in] this Chemical model [in] env_states Environmental state array (one element per grid cell to solve simultaneously)
Definition at line 982 of file camp_core.F90.
◆ new_state_one_cell()
|
private |
Get a model state variable based on the this set of model data This is also called for multi-cell systems when no env_state_t array is passed.
- Returns
- New model state
- Parameters
-
[in] this Chemical model [in] env_state Environmental state array (one element per grid cell to solve simultaneously)
Definition at line 1005 of file camp_core.F90.

◆ pack_size()
| integer(kind=i_kind) function camp_camp_core::pack_size | ( | class(camp_core_t), intent(in) | this, |
| integer, intent(in), optional | comm | ||
| ) |
Determine the size of a binary required to pack the mechanism.
- Parameters
-
[in] this Chemical model [in] comm MPI communicator
Definition at line 1417 of file camp_core.F90.


◆ rxn_update_data()
|
private |
Update data associated with a reaction. This function should be called when reaction parameters need updated from the host model. For example, this function can be called to update photolysis rates from a host model's photolysis module.
- Parameters
-
[in] this Chemical model [in] update_data Update data
Definition at line 1313 of file camp_core.F90.
◆ solve()
|
private |
Integrate the chemical mechanism.
- Parameters
-
[in] this Chemical model [in,out] camp_state Current model state [in] time_step Time step over which to integrate (s) [in] rxn_phase Phase to solve - gas, aerosol, or both (default) Use parameters in camp_rxn_data to specify phase: GAS_RXN, AERO_RXN, GAS_AERO_RXN [in,out] solver_stats Return solver statistics to the host model
Definition at line 1352 of file camp_core.F90.

◆ solver_initialize()
|
private |
Initialize the solver.
- Parameters
-
[in,out] this Chemical model
Definition at line 1134 of file camp_core.F90.

◆ spec_state_id()
|
private |
Get the id of a species on the state array by its unique name.
- Returns
- Flag indicating whether the species was found
- Parameters
-
[in] this CAMP core [in] spec_name Species unique name [in,out] state_id Species state id
Definition at line 1104 of file camp_core.F90.
◆ state_size()
|
private |
Get the size of the state array.
- Returns
- State size
- Parameters
-
[in] this Chemical model
Definition at line 1033 of file camp_core.F90.

◆ state_size_per_cell()
|
private |
Get the size of the state array for each grid cell.
- Returns
- State size
- Parameters
-
[in] this Chemical model
Definition at line 1051 of file camp_core.F90.
◆ sub_model_update_data()
|
private |
Update data associated with a sub-model. This function should be called when sub-model parameters need updated from the host model.
- Parameters
-
[in] this Chemical model [in] update_data Update data
Definition at line 1333 of file camp_core.F90.
◆ unique_names()
|
private |
Get an array of unique names for all species on the state array.
The order of this array is the same as the state array for one grid cell
- Returns
- Array of unique species names
- Parameters
-
[in] this CAMP core
Definition at line 1071 of file camp_core.F90.