UNIFAC activity coefficient calculation. More...
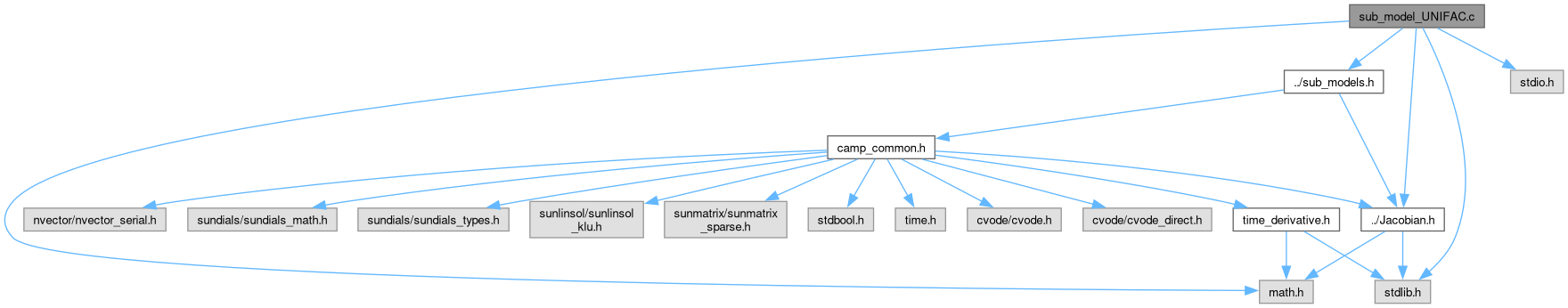
Go to the source code of this file.
Macros | |
| #define | TEMPERATURE_K_ env_data[0] |
| #define | PRESSURE_PA_ env_data[1] |
| #define | NUM_UNIQUE_PHASE_ (int_data[0]) |
| #define | NUM_GROUP_ (int_data[1]) |
| #define | TOTAL_INT_PROP_ (int_data[2]) |
| #define | TOTAL_FLOAT_PROP_ (int_data[3]) |
| #define | NUM_INT_PROP_ 4 |
| #define | NUM_FLOAT_PROP_ 0 |
| #define | PHASE_INT_LOC_(p) (int_data[NUM_INT_PROP_ + p] - 1) |
| #define | PHASE_FLOAT_LOC_(p) (int_data[NUM_INT_PROP_ + NUM_UNIQUE_PHASE_ + p] - 1) |
| #define | PHASE_ENV_LOC_(p) (int_data[NUM_INT_PROP_ + 2 * NUM_UNIQUE_PHASE_ + p] - 1) |
| #define | NUM_PHASE_INSTANCE_(p) (int_data[PHASE_INT_LOC_(p)]) |
| #define | NUM_SPEC_(p) (int_data[PHASE_INT_LOC_(p) + 1]) |
| #define | PHASE_INST_ID_(p, c) (int_data[PHASE_INT_LOC_(p) + 2 + c] - 1) |
| #define | SPEC_ID_(p, i) (int_data[PHASE_INT_LOC_(p) + 2 + NUM_PHASE_INSTANCE_(p) + i]) |
| #define | GAMMA_ID_(p, i) (int_data[PHASE_INT_LOC_(p) + 2 + NUM_PHASE_INSTANCE_(p) + NUM_SPEC_(p) + i]) |
| #define | JAC_ID_(p, c, j, i) |
| #define | V_IK_(p, i, k) |
| #define | Q_K_(k) (float_data[k]) |
| #define | R_K_(k) (float_data[NUM_GROUP_ + k]) |
| #define | X_K_(m) (float_data[2 * NUM_GROUP_ + m]) |
| #define | DTHETA_M_DC_I_(m) (float_data[3 * NUM_GROUP_ + m]) |
| #define | XI_M_(m) (float_data[4 * NUM_GROUP_ + m]) |
| #define | LN_GAMMA_K_(m) (float_data[5 * NUM_GROUP_ + m]) |
| #define | A_MN_(m, n) (float_data[(m + 6) * NUM_GROUP_ + n]) |
| #define | R_I_(p, i) (float_data[PHASE_FLOAT_LOC_(p) + i]) |
| #define | Q_I_(p, i) (float_data[PHASE_FLOAT_LOC_(p) + NUM_SPEC_(p) + i]) |
| #define | L_I_(p, i) (float_data[PHASE_FLOAT_LOC_(p) + 2 * NUM_SPEC_(p) + i]) |
| #define | MW_I_(p, i) (float_data[PHASE_FLOAT_LOC_(p) + 3 * NUM_SPEC_(p) + i]) |
| #define | X_I_(p, i) (float_data[PHASE_FLOAT_LOC_(p) + 4 * NUM_SPEC_(p) + i]) |
| #define | THETA_M_(m) (sub_model_env_data[m]) |
| #define | PSI_MN_(m, n) (sub_model_env_data[(m + 1) * NUM_GROUP_ + n]) |
| #define | LN_GAMMA_IK_(p, i, k) (sub_model_env_data[PHASE_ENV_LOC_(p) + i * NUM_GROUP_ + k]) |
| #define | INT_DATA_SIZE_ (TOTAL_INT_PROP_) |
| #define | FLOAT_DATA_SIZE_ (TOTAL_FLOAT_PROP_) |
Functions | |
| void | sub_model_UNIFAC_get_used_jac_elem (int *sub_model_int_data, double *sub_model_float_data, Jacobian *jac) |
| Get the Jacobian elements used for a particular row of the matrix. More... | |
| void | sub_model_UNIFAC_update_ids (int *sub_model_int_data, double *sub_model_float_data, int *deriv_ids, Jacobian jac) |
| Update stored ids for elements used within a row of the Jacobian matrix. More... | |
| void | sub_model_UNIFAC_update_env_state (int *sub_model_int_data, double *sub_model_float_data, double *sub_model_env_data, ModelData *model_data) |
| Update sub-model data for new environmental conditions. More... | |
| void | sub_model_UNIFAC_calculate (int *sub_model_int_data, double *sub_model_float_data, double *sub_model_env_data, ModelData *model_data) |
| Perform the sub-model calculations for the current model state. More... | |
| void | sub_model_UNIFAC_get_jac_contrib (int *sub_model_int_data, double *sub_model_float_data, double *sub_model_env_data, ModelData *model_data, realtype *J, double time_step) |
| Add contributions to the Jacobian from derivates calculated using the output of this sub model. More... | |
| void | sub_model_UNIFAC_print (int *sub_model_int_data, double *sub_model_float_data) |
| Print the sub model data. More... | |
Detailed Description
UNIFAC activity coefficient calculation.
For more info see the camp_sub_model_UNIFAC module
Equation references are to Marcolli and Peter, ACP 5(2), 1501-1527, 2005.
Definition in file sub_model_UNIFAC.c.
Macro Definition Documentation
◆ A_MN_
| #define A_MN_ | ( | m, | |
| n | |||
| ) | (float_data[(m + 6) * NUM_GROUP_ + n]) |
Definition at line 58 of file sub_model_UNIFAC.c.
◆ DTHETA_M_DC_I_
| #define DTHETA_M_DC_I_ | ( | m | ) | (float_data[3 * NUM_GROUP_ + m]) |
Definition at line 55 of file sub_model_UNIFAC.c.
◆ FLOAT_DATA_SIZE_
| #define FLOAT_DATA_SIZE_ (TOTAL_FLOAT_PROP_) |
Definition at line 71 of file sub_model_UNIFAC.c.
◆ GAMMA_ID_
| #define GAMMA_ID_ | ( | p, | |
| i | |||
| ) | (int_data[PHASE_INT_LOC_(p) + 2 + NUM_PHASE_INSTANCE_(p) + NUM_SPEC_(p) + i]) |
Definition at line 42 of file sub_model_UNIFAC.c.
◆ INT_DATA_SIZE_
| #define INT_DATA_SIZE_ (TOTAL_INT_PROP_) |
Definition at line 70 of file sub_model_UNIFAC.c.
◆ JAC_ID_
| #define JAC_ID_ | ( | p, | |
| c, | |||
| j, | |||
| i | |||
| ) |
Definition at line 44 of file sub_model_UNIFAC.c.
◆ L_I_
| #define L_I_ | ( | p, | |
| i | |||
| ) | (float_data[PHASE_FLOAT_LOC_(p) + 2 * NUM_SPEC_(p) + i]) |
Definition at line 61 of file sub_model_UNIFAC.c.
◆ LN_GAMMA_IK_
| #define LN_GAMMA_IK_ | ( | p, | |
| i, | |||
| k | |||
| ) | (sub_model_env_data[PHASE_ENV_LOC_(p) + i * NUM_GROUP_ + k]) |
Definition at line 67 of file sub_model_UNIFAC.c.
◆ LN_GAMMA_K_
| #define LN_GAMMA_K_ | ( | m | ) | (float_data[5 * NUM_GROUP_ + m]) |
Definition at line 57 of file sub_model_UNIFAC.c.
◆ MW_I_
| #define MW_I_ | ( | p, | |
| i | |||
| ) | (float_data[PHASE_FLOAT_LOC_(p) + 3 * NUM_SPEC_(p) + i]) |
Definition at line 62 of file sub_model_UNIFAC.c.
◆ NUM_FLOAT_PROP_
| #define NUM_FLOAT_PROP_ 0 |
Definition at line 31 of file sub_model_UNIFAC.c.
◆ NUM_GROUP_
| #define NUM_GROUP_ (int_data[1]) |
Definition at line 27 of file sub_model_UNIFAC.c.
◆ NUM_INT_PROP_
| #define NUM_INT_PROP_ 4 |
Definition at line 30 of file sub_model_UNIFAC.c.
◆ NUM_PHASE_INSTANCE_
| #define NUM_PHASE_INSTANCE_ | ( | p | ) | (int_data[PHASE_INT_LOC_(p)]) |
Definition at line 37 of file sub_model_UNIFAC.c.
◆ NUM_SPEC_
| #define NUM_SPEC_ | ( | p | ) | (int_data[PHASE_INT_LOC_(p) + 1]) |
Definition at line 38 of file sub_model_UNIFAC.c.
◆ NUM_UNIQUE_PHASE_
| #define NUM_UNIQUE_PHASE_ (int_data[0]) |
Definition at line 26 of file sub_model_UNIFAC.c.
◆ PHASE_ENV_LOC_
| #define PHASE_ENV_LOC_ | ( | p | ) | (int_data[NUM_INT_PROP_ + 2 * NUM_UNIQUE_PHASE_ + p] - 1) |
Definition at line 35 of file sub_model_UNIFAC.c.
◆ PHASE_FLOAT_LOC_
| #define PHASE_FLOAT_LOC_ | ( | p | ) | (int_data[NUM_INT_PROP_ + NUM_UNIQUE_PHASE_ + p] - 1) |
Definition at line 33 of file sub_model_UNIFAC.c.
◆ PHASE_INST_ID_
| #define PHASE_INST_ID_ | ( | p, | |
| c | |||
| ) | (int_data[PHASE_INT_LOC_(p) + 2 + c] - 1) |
Definition at line 39 of file sub_model_UNIFAC.c.
◆ PHASE_INT_LOC_
| #define PHASE_INT_LOC_ | ( | p | ) | (int_data[NUM_INT_PROP_ + p] - 1) |
Definition at line 32 of file sub_model_UNIFAC.c.
◆ PRESSURE_PA_
| #define PRESSURE_PA_ env_data[1] |
Definition at line 24 of file sub_model_UNIFAC.c.
◆ PSI_MN_
| #define PSI_MN_ | ( | m, | |
| n | |||
| ) | (sub_model_env_data[(m + 1) * NUM_GROUP_ + n]) |
Definition at line 66 of file sub_model_UNIFAC.c.
◆ Q_I_
| #define Q_I_ | ( | p, | |
| i | |||
| ) | (float_data[PHASE_FLOAT_LOC_(p) + NUM_SPEC_(p) + i]) |
Definition at line 60 of file sub_model_UNIFAC.c.
◆ Q_K_
| #define Q_K_ | ( | k | ) | (float_data[k]) |
Definition at line 52 of file sub_model_UNIFAC.c.
◆ R_I_
| #define R_I_ | ( | p, | |
| i | |||
| ) | (float_data[PHASE_FLOAT_LOC_(p) + i]) |
Definition at line 59 of file sub_model_UNIFAC.c.
◆ R_K_
| #define R_K_ | ( | k | ) | (float_data[NUM_GROUP_ + k]) |
Definition at line 53 of file sub_model_UNIFAC.c.
◆ SPEC_ID_
| #define SPEC_ID_ | ( | p, | |
| i | |||
| ) | (int_data[PHASE_INT_LOC_(p) + 2 + NUM_PHASE_INSTANCE_(p) + i]) |
Definition at line 40 of file sub_model_UNIFAC.c.
◆ TEMPERATURE_K_
| #define TEMPERATURE_K_ env_data[0] |
Definition at line 23 of file sub_model_UNIFAC.c.
◆ THETA_M_
| #define THETA_M_ | ( | m | ) | (sub_model_env_data[m]) |
Definition at line 65 of file sub_model_UNIFAC.c.
◆ TOTAL_FLOAT_PROP_
| #define TOTAL_FLOAT_PROP_ (int_data[3]) |
Definition at line 29 of file sub_model_UNIFAC.c.
◆ TOTAL_INT_PROP_
| #define TOTAL_INT_PROP_ (int_data[2]) |
Definition at line 28 of file sub_model_UNIFAC.c.
◆ V_IK_
| #define V_IK_ | ( | p, | |
| i, | |||
| k | |||
| ) |
Definition at line 47 of file sub_model_UNIFAC.c.
◆ X_I_
| #define X_I_ | ( | p, | |
| i | |||
| ) | (float_data[PHASE_FLOAT_LOC_(p) + 4 * NUM_SPEC_(p) + i]) |
Definition at line 63 of file sub_model_UNIFAC.c.
◆ X_K_
| #define X_K_ | ( | m | ) | (float_data[2 * NUM_GROUP_ + m]) |
Definition at line 54 of file sub_model_UNIFAC.c.
◆ XI_M_
| #define XI_M_ | ( | m | ) | (float_data[4 * NUM_GROUP_ + m]) |
Definition at line 56 of file sub_model_UNIFAC.c.
Function Documentation
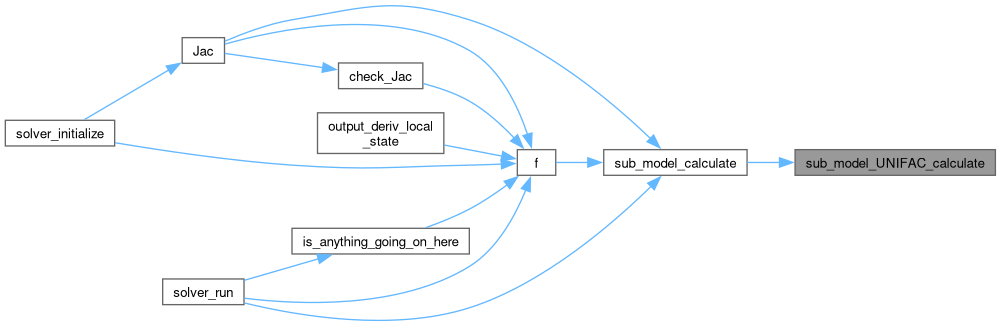
◆ sub_model_UNIFAC_calculate()
| void sub_model_UNIFAC_calculate | ( | int * | sub_model_int_data, |
| double * | sub_model_float_data, | ||
| double * | sub_model_env_data, | ||
| ModelData * | model_data | ||
| ) |
Perform the sub-model calculations for the current model state.
These calculations are based on equations (1)–(9) in [6]. Variable names used in the derivation of the Jacobian equations are adopted here for consistency. Specifically, these are:
\[ \begin{align*} \sigma & \equiv \displaystyle\sum_k r_k x_k, \\ \tau & \equiv \displaystyle\sum_k q_k x_k, \\ \mu & \equiv \displaystyle\sum_j x_j l_j, \\ c_{xX} & = \frac{1}{\displaystyle\sum_j \displaystyle\sum_i v_j^{(i)} x_i}, \textrm{ and} \\ \Pi & \equiv \displaystyle\sum_n Q_n X_n. \\ \end{align*} \]
Using these variables and subscript \(j\) for the species whose activity is being calculated for easy comparison with the Jacobian calculations, equations (1)–(9) in [6] become:
\[ \begin{align*} \ln{\gamma_j} & = \ln{\gamma_j^C} + \ln{\gamma_j^R} & (1) \\ \alpha_w & = \gamma_w x_w & (2) \\ \ln{\gamma_j^C} & = \ln{\frac{\Phi_j}{x_j}} + \frac{z}{2}q_j\ln{\frac{\Theta_j}{\Phi_j}} + l_j - \frac{\Phi_j}{x_j}\mu & (3) \\ \Phi_j & = \frac{r_j x_j}{\sigma}; \qquad \Theta_j = \frac{q_j x_j}{\tau} & (4) \\ l_j & = \frac{z}{2}\left(r_j - q_j\right) - r_j + 1 & (5) \\ r_j & = \displaystyle\sum_k v_k^{(j)}R_k; \qquad q_j = \displaystyle\sum_k v_k^{(j)}Q_k & (6) \\ \ln{\gamma_j^R} & = \displaystyle\sum_k v_k^{(j)} \left[ \ln{\Gamma_k} - \ln{\Gamma_k^{(j)}}\right] & (7) \\ \ln{\Gamma_k} & = Q_k \left[ 1 - \ln{\Xi_k} - \displaystyle\sum_m\frac{\Theta_m\Phi_{km}}{\Xi_m} \right] & (8) \\ \Theta_m & = \frac{Q_m X_m}{\Pi}; \qquad \Psi_{mn} = \mathrm{exp}\left[\frac{-a_{mn}}{T}\right] & (9) \\ \end{align*} \]
- Parameters
-
sub_model_int_data Pointer to the sub model integer data sub_model_float_data Pointer to the sub model floating-point data sub_model_env_data Pointer to the sub model environment-dependent data model_data Pointer to the model data including the current state and environmental conditions
Definition at line 226 of file sub_model_UNIFAC.c.
◆ sub_model_UNIFAC_get_jac_contrib()
| void sub_model_UNIFAC_get_jac_contrib | ( | int * | sub_model_int_data, |
| double * | sub_model_float_data, | ||
| double * | sub_model_env_data, | ||
| ModelData * | model_data, | ||
| realtype * | J, | ||
| double | time_step | ||
| ) |
Add contributions to the Jacobian from derivates calculated using the output of this sub model.
This derivation starts from equations (1)–(9) in [6]. The mole fraction of species \(i\) is calculated as:
\[ x_i = \frac{m_i}{m_T}, \]
where \(m_T = \displaystyle\sum_j m_j\), \(m_i = \frac{c_i}{\mathrm{MW}_i}\) is the number in moles of species \(i\), \(c_i\) is its mass concentration ( \(\mathrm{ug} \: \mathrm{m}^{-3}\); the state variable), and \(\mathrm{MW}_i\) is its molecular weight ( \(\mathrm{ug} \: \mathrm{mol}^{-1}\)). Thus,
\[ \begin{align*} \frac{\partial x_i}{\partial c_i} & = \frac{(m_T-m_i)}{\mathrm{MW}_i m_T^2}, \textrm{ and} \\ \frac{\partial x_j}{\partial c_i} & = -\frac{m_j}{\mathrm{MW}_i m_T^2} \quad \text{for } i\neq j. \\ \end{align*} \]
The partial derivative of \(\Phi_j\) (Eq. 4) with respect to \(c_i\) is derived as follows:
\[ \begin{align*} \frac{\partial r_i x_i}{\partial c_i} & = r_i \frac{(m_T-m_i)}{\mathrm{MW}_i m_T^2}, \\ \frac{\partial r_j x_j}{\partial c_i} & = - r_j \frac{m_j}{\mathrm{MW}_i m_T^2} \quad \text{for } i\neq j, \\ \frac{\partial \displaystyle\sum_j r_j x_j}{\partial c_i} & = \frac{1}{\mathrm{MW}_i m_T}\left[r_i - \displaystyle\sum_j r_j x_j\right], \\ \sigma & \equiv \displaystyle\sum_k r_k x_k, \\ \frac{\partial \Phi_j}{\partial c_i} & = \frac{r_j}{\mathrm{MW}_i m_T \sigma^2} \left(\sigma^{\prime} - x_j r_i \right) , \qquad \sigma^{\prime} = \begin{cases} \sigma & \quad \text{if } i=j \\ 0 & \quad \text{if } i\neq j \end{cases} \\ \end{align*} \]
Similarly, the partial derivative of \(\Theta_j\) (Eq. 4) with respect to \(c_i\) is:
\[ \begin{align*} \tau & \equiv \displaystyle\sum_k q_k x_k, \\ \frac{\partial \Theta_j}{\partial c_i} & = \frac{q_j}{\mathrm{MW}_i m_T \tau^2} \left(\tau^{\prime} - x_j q_i \right) , \qquad \tau^{\prime} = \begin{cases} \tau & \quad \text{if } i=j \\ 0 & \quad \text{if } i\neq j \end{cases} \\ \end{align*} \]
From Eqs 5 and 6,
\[ \frac{\partial r_j}{\partial c_i} = \frac{\partial q_j}{\partial c_i} = \frac{\partial l_j}{\partial c_i} = 0. \]
For the last term in Eq. 3,
\[ \begin{align*} \mu & \equiv \displaystyle\sum_j x_j l_j, \\ \frac{\partial \mu}{\partial c_i} & = \frac{1}{\mathrm{MW}_i m_T} \left( l_i - \mu \right). \\ \end{align*} \]
The partial derivative of the full combinatorial term (Eq. 3) is:
\[ \begin{align*} \frac{\partial \ln{\gamma^C_j}}{\partial c_i} & = \frac{x_j}{\Phi_j}\frac{\left( \frac{\partial \Phi_j}{\partial c_i} x_j - \Phi_j\frac{\partial x_j}{\partial c_i} \right)}{x_j^2} \\ & \quad + \frac{z}{2}q_j\frac{\Phi_j}{\Theta_j}\frac{\left( \frac{\partial \Theta_j}{\partial c_i} \Phi_j - \Theta_j\frac{\partial \Phi_j}{\partial c_i}\right)}{\Phi_j^2} \\ & \quad - \frac{\left[ \left( \frac{\partial \Phi_j}{\partial c_i} \mu + \Phi_j \frac{\partial \mu}{\partial c_i} \right) x_j - \Phi_j \mu \frac{\partial x_j}{\partial c_i} \right]}{x_j^2}. \end{align*} \]
After some rearranging, this becomes:
\[ \begin{align*} \frac{\partial \ln{\gamma^C_j}}{\partial c_i} & = \frac{1}{\Phi_j}\frac{\partial \Phi_j}{\partial c_i} - \frac{1}{x_j}\frac{\partial x_j}{\partial c_i} \\ & \quad + \frac{z}{2}q_j\left( \frac{1}{\Theta_j}\frac{\partial \Theta_j}{\partial c_i} - \frac{1}{\Phi_j}\frac{\partial \Phi_j}{\partial c_i}\right) \\ & \quad - \frac{\partial \Phi_j}{\partial c_i}\frac{\mu}{x_j} - \frac{\Phi_j}{x_j}\frac{\partial \mu}{\partial c_i} + \frac{\Phi_j \mu}{x_j^2}\frac{\partial x_j}{\partial c_i} \end{align*} \]
As \(\sigma\), \(\tau\), and \(\mu\) are independent of species \(i\), these can be calculated outside the loop over the independent species. Moving to the residual term (Eq. 7), the mole fraction ( \(X_p\)) of group \(p\) in the mixture is related to the mole fraction ( \(x_j\)) of species \(j\) according to:
\[ X_p = c_{xX} \omega_p, \textrm{ where } \omega_p \equiv \displaystyle\sum_j v_p^{(j)} x_j, \\ \]
and \(c_{xX}\) is a conversion factor accounting for the difference in total species and total group number concentrations:
\[ c_{xX} = \frac{1}{\displaystyle\sum_p \omega_p}. \]
Partial derivatives of the group interaction terms in Eq 9, \(\Theta_m\) and \(\Psi_{mn}\), with respect to \(c_i\) are derived as follows:
\[ \begin{align*} \frac{\partial c_{xX}}{\partial c_i} & = - c_{xX}^2 \displaystyle\sum_p \displaystyle\sum_j v_p^{(j)} \frac{\partial x_j}{\partial c_i} = - \frac{c_{xX}^2}{\mathrm{MW}_im_T} \left( \displaystyle\sum_p v_p^{(i)} - \displaystyle\sum_p \omega_p \right), \\ & = \frac{c_{xX}}{\mathrm{MW}_im_T} \left( 1 - c_{xX} \displaystyle\sum_p v_p^{(i)} \right), \\ \frac{\partial X_n}{\partial c_i} & = c_{xX} \displaystyle\sum_j v_n^{(j)} \frac{\partial x_j}{\partial c_i} + \frac{\partial c_{xX}}{\partial c_i} \omega_n, \\ & = \frac{c_{xX}}{\mathrm{MW}_im_T} \left( v_n^{(i)} - \omega_n \right) + \frac{c_{xX}}{\mathrm{MW}_im_T} \left( \omega_n - c_{xX}\omega_n \displaystyle\sum_p v_p^{(i)} \right), \\ & = \frac{c_{xX}}{\mathrm{MW}_im_T} \left( v_n^{(i)} - X_n \displaystyle\sum_p v_p^{(i)} \right), \\ \Pi & \equiv \displaystyle\sum_n Q_n X_n, \\ \frac{\partial \Pi}{\partial c_i} & = \displaystyle\sum_n Q_n \frac{c_{xX}}{\mathrm{MW}_i m_T}\left( v_n^{(i)} - X_n \displaystyle\sum_p v_p^{(i)} \right), \\ & = \frac{c_{xX}}{\mathrm{MW}_i m_T} \left( \displaystyle\sum_n Q_n v_n^{(i)} - \Pi \displaystyle\sum_p v_p^{(i)} \right), \\ \frac{\partial \Theta_m}{\partial c_i} & = \frac{ \left(Q_m\frac{\partial X_m}{\partial c_i} \Pi - Q_m X_m \frac{\partial \Pi}{\partial c_i} \right)}{\Pi^2} , \\ & = \frac{c_{xX} Q_m}{\mathrm{MW}_im_T\Pi^2} \left[ \left( \Pi v_m^{(i)} - \Pi X_m \displaystyle\sum_p v_p^{(i)} \right) - \left( X_m \displaystyle\sum_n Q_n v_n^{(i)} - X_m \Pi \displaystyle\sum_p v_p^{(i)} \right) \right], \\ & = \frac{c_{xX} Q_m}{\mathrm{MW}_im_T\Pi^2} \left[ \Pi v_m^{(i)} - X_m\displaystyle\sum_n Q_n v_n^{(i)}\right], \textrm{ and} \\ \frac{\partial \Psi_{mn}}{\partial c_i} & = 0 \end{align*} \]
The partial derivative of the group residual activity coefficient (Eq. 8) with respect to \(c_i\) is:
\[ \begin{align*} \frac{\partial \ln{\Gamma_k}}{\partial c_i} & = Q_k \left[ - \frac{1}{\displaystyle\sum_m\Theta_m\Psi_{mk}} \displaystyle\sum_m\frac{\partial\Theta_m}{\partial c_i}\Psi_{mk} \\ \quad - \displaystyle\sum_m\left( \frac{\partial\Theta_m}{\partial c_i}\Psi_{km} \displaystyle\sum_n\Theta_n\Psi_{nm} - \Theta_m\Psi_{km}\displaystyle\sum_n \frac{\partial\Theta_n}{\partial c_i} \Psi_{nm}\right)/\left( \displaystyle\sum_n\Theta_n\Psi_{nm}\right)^2 \right] \end{align*} \]
After some rearranging, this becomes:
\[ \begin{align*} \Xi_m & \equiv \displaystyle\sum_n\Theta_n\Psi_{nm}, \\ \frac{\partial \ln{\Gamma_k}}{\partial c_i} & = - Q_k \displaystyle\sum_m \left( \frac{\Psi_{mk}}{\Xi_k}\frac{\partial\Theta_m}{\partial c_i} + \frac{\Psi_{km}}{\Xi_m} \frac{\partial\Theta_m}{\partial c_i} - \frac{\Theta_m\Psi_{km}} {\Xi_m^2} \displaystyle\sum_n\frac{\partial\Theta_n}{\partial c_i}\Psi_{nm} \right) \end{align*} \]
The left side of the three bracketed terms in the above equation are independent of species \(i\) and can be calculated outside of the loop over the independent species. The partial derivative of the full residual term with respect to \(c_i\) is:
\[ \begin{align*} \frac{\partial \ln{\Gamma_k^{(j)}}}{\partial c_i} & = 0 \\ \frac{\partial \ln{\gamma_j^R}}{\partial c_i} & = \displaystyle\sum_k v_k^{(j)} \frac{\partial \ln{\Gamma_k}}{\partial c_i} \end{align*} \]
The overall equation for the partial derivative of \(\gamma_j\) with respect to species \(i\) is:
\[ \frac{\partial\gamma_j}{\partial c_i} = \gamma_j \left( \frac{\partial\ln{\gamma_j^C}}{\partial c_i} + \frac{\partial\ln{\gamma_j^R}}{\partial c_i} \right) \]
- Parameters
-
sub_model_int_data Pointer to the sub model integer data sub_model_float_data Pointer to the sub model floating-point data sub_model_env_data Pointer to the sub model environment-dependent data model_data Pointer to the model data J Jacobian to be calculated time_step Current time step [s]
Definition at line 575 of file sub_model_UNIFAC.c.

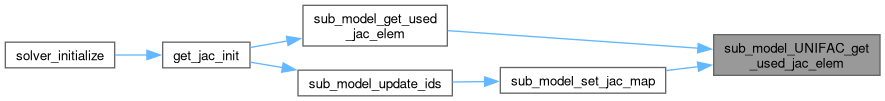
◆ sub_model_UNIFAC_get_used_jac_elem()
| void sub_model_UNIFAC_get_used_jac_elem | ( | int * | sub_model_int_data, |
| double * | sub_model_float_data, | ||
| Jacobian * | jac | ||
| ) |
Get the Jacobian elements used for a particular row of the matrix.
- Parameters
-
sub_model_int_data Pointer to the sub model integer data sub_model_float_data Pointer to the sub model floating-point data jac Jacobian
Definition at line 82 of file sub_model_UNIFAC.c.

◆ sub_model_UNIFAC_print()
| void sub_model_UNIFAC_print | ( | int * | sub_model_int_data, |
| double * | sub_model_float_data | ||
| ) |
Print the sub model data.
- Parameters
-
sub_model_int_data Pointer to the sub model integer data sub_model_float_data Pointer to the sub model floating-point data
Definition at line 771 of file sub_model_UNIFAC.c.

◆ sub_model_UNIFAC_update_env_state()
| void sub_model_UNIFAC_update_env_state | ( | int * | sub_model_int_data, |
| double * | sub_model_float_data, | ||
| double * | sub_model_env_data, | ||
| ModelData * | model_data | ||
| ) |
Update sub-model data for new environmental conditions.
- Parameters
-
sub_model_int_data Pointer to the sub model integer data sub_model_float_data Pointer to the sub model floating-point data sub_model_env_data Pointer to the sub model environment-dependent data model_data Pointer to the model data
Definition at line 129 of file sub_model_UNIFAC.c.

◆ sub_model_UNIFAC_update_ids()
| void sub_model_UNIFAC_update_ids | ( | int * | sub_model_int_data, |
| double * | sub_model_float_data, | ||
| int * | deriv_ids, | ||
| Jacobian | jac | ||
| ) |
Update stored ids for elements used within a row of the Jacobian matrix.
- Parameters
-
sub_model_int_data Pointer to the sub model integer data sub_model_float_data Pointer to the sub model floating-point data deriv_ids Indices for state array variables on the solver state array jac Jacobian
Definition at line 106 of file sub_model_UNIFAC.c.

